Transcription in Prokaryotes
After the DNA is replicated during cell division, the cell needs them to convert to RNA for forming proteins. The process of converting the DNA sequence of a gene to an RNA molecule is called transcription. The enzyme carrying out transcription is called RNA polymerase.
Transcription is almost similar in prokaryotes and eukaryotes. Here, we will discuss transcription by describing the process in the bacterium Escherichia coli.
Where does Transcription Occur in Prokaryotes
Since E. coli is single-celled and does not have membrane-enclosed nuclei, transcription occurs in the cell’s cytoplasm.
Structure of RNA Polymerase
It is the enzyme that carries transcription. Prokaryotes utilize the same RNA polymerase to transcribe their genes. In E. coli, the polymerase consists of five polypeptide subunits – two α, one β, one β’, and one ω and σ subunit (α2ββ’ωσ).
The polymerase with all five subunits is called a holoenzyme. These subunits assemble before a gene is to be transcribed and disassemble once the transcription is complete. When the σ subunit dissociates, it forms the core enzyme.
Stages of Transcription
For transcription, the DNA double helix must unwind partially in the region of mRNA synthesis. The region is called the transcription bubble.
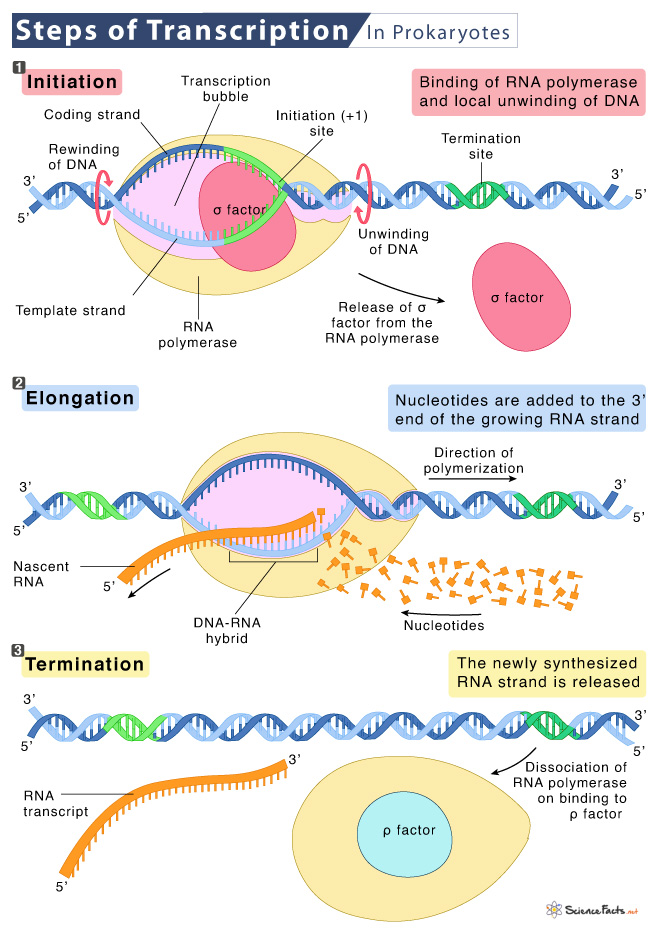
The DNA strand that RNA polymerase uses to form RNA is the template strand, whereas the other strand is the non-template strand or the coding strand. The mRNA is complementary to the template strand and is almost identical to the coding strand.
The nucleotide pair in the DNA double helix from which the first five mRNA nucleotide is transcribed is called the +1 site, or the initiation site. The region above the initiation site is called upstream and is denoted by (-). In contrast, those following the initiation site downstream are called downstream and are denoted by (+).
1. Initiation
It is the first step of transcription. The RNA polymerase holoenzyme can recognize and bind to specific regions in the DNA (consensus sequence) upstream of the initiation site at the -10 and -35 regions. The -10 consensus has the sequence TATAAT and -35, TTGACA. These two promoter consensus sequences are similar across all promoters and various bacterial species.
Once bound to the promoter, the σ subunit dissociates to form the core enzyme.
2. Elongation
This phase begins with releasing the σ subunit from the RNA polymerase. The dissociation of the σ subunit allows the core enzyme to proceed along the DNA template, synthesizing mRNA in the 5′ to 3′ direction at approximately 40 nucleotides per second rate.
As elongation proceeds from the 5’ to 3’ direction by adding nucleotides (NTPs) to the 3’ end of the nascent RNA, the DNA continuously unwinds ahead of the core enzyme and rewinds behind it.
The bond between the DNA and the nascent RNA being unstable, the RNA polymerase acts as a linker between the two strands to stabilize the duplex. It allows RNA polymerase to continue synthesizing the RNA without interruptions.
3. Termination
Once the transcription is complete, the RNA polymerase must dissociate from the template, releasing the newly formed RNA molecule.
Depending on the gene, there are two types of termination signals.
a. Rho-dependent termination is controlled by the rho protein, which moves behind the RNA polymerase on the growing mRNA chain. At the end of the gene, the polymerase encounters a run of G nucleotides on the template and stops polymerizing the strand. As a result, the RNA polymerase collides with the rho protein, releasing the mRNA from the transcription bubble.
b. Rho-independent termination depends on specific sequences on the DNA strand. When the polymerase approaches the end of a gene, it encounters a region rich in C–G nucleotides. The mRNA then folds back on itself, allowing the complementary G-C nucleotides to bind, forming a stable hairpin structure. It causes the polymerase to stall transcription of a region rich in A–T nucleotides.
The complementary U-A region of the transcript forms only a weak interaction with the template DNA. Together with stalled RNA polymerase, it destabilizes the core enzyme complex to break away from the new mRNA transcript.
What Happens to the RNA Transcript after Transcription
Since transcription and translation occur concurrently in prokaryotes, the mRNA transcript is used to make numerous copies of proteins even before the end of transcription. More than one copy of polymerase can transcribe a single bacterial gene at a particular time. It helps to produce a high number of RNA molecules necessary to meet the protein requirement of the cell.
FAQs
Ans. No, prokaryotes do not have transcription factors.
-
References
Article was last reviewed on Thursday, February 2, 2023