Trp Operon
A trp operon is a group of genes in prokaryotic DNA that encodes proteins involved in tryptophan synthesis, an essential amino acid in living organisms.
When tryptophan levels are low in the bacterial cell, the trp operon is activated, promoting tryptophan synthesis. Conversely, when tryptophan is abundant, the trp operon is repressed, conserving energy by reducing the production of tryptophan. The trp operon is thus a repressible system.
Components of the Trp Operon
The trp operon consists of five key elements:
1. Repressor Protein (trpR): TrpR is a regulatory protein synthesized by a separate gene. It acts as a switch for the operon. During the absence of tryptophan, the repressor protein cannot bind to the operator region.
2. Operator Region: This is a DNA segment located before the structural genes of the operon. It serves as the binding site for the repressor protein. When the repressor is bound to the operator, it prevents RNA polymerase from transcribing the structural genes.
3. Promoter Region: Found adjacent to the operator, the promoter is the site where RNA polymerase binds to initiate transcription of the trp genes.
4. Structural Genes: The trp operon contains a series of genes that encodes enzymes involved in tryptophan biosynthesis. These genes include trpE, trpD, trpC, trpB, and trpA.
- trpE: Codes for the enzyme anthranilate synthase, which catalyzes the conversion of chorismate to anthranilate.
- trpD: Codes for indole-3-glycerol phosphate synthase, which catalyzes the conversion of indole-3-glycerol phosphate to indole and glyceraldehyde-3-phosphate.
- trpC: Codes for indole-3-glycerol phosphate lyase, which catalyzes the conversion of indole-3-glycerol phosphate to indole.
- trpB: Codes for tryptophan synthase beta subunit, which catalyzes the conversion of indole and serine to tryptophan.
- trpA: Codes for tryptophan synthase alpha subunit, which catalyzes the final step in tryptophan biosynthesis, combining indole and serine to produce tryptophane.
5. Attenuator Region: In some bacterial species, an attenuator region is found in the leader sequence of the mRNA transcribed from the Trp Operon. The attenuator region plays a role in fine-tuning the regulation of the operon. It is involved in the concept of attenuation. This mechanism can influence whether transcription proceeds to the structural genes or is terminated prematurely.
The attenuator region contains specific sequences that can form alternative secondary structures based on the availability of tryptophan in the cell.
How Does the Trp Operon Work
The regulation of the trp operon is primarily controlled at the transcriptional level. The presence or absence of tryptophan in the bacterial cell determines the operon’s activity.
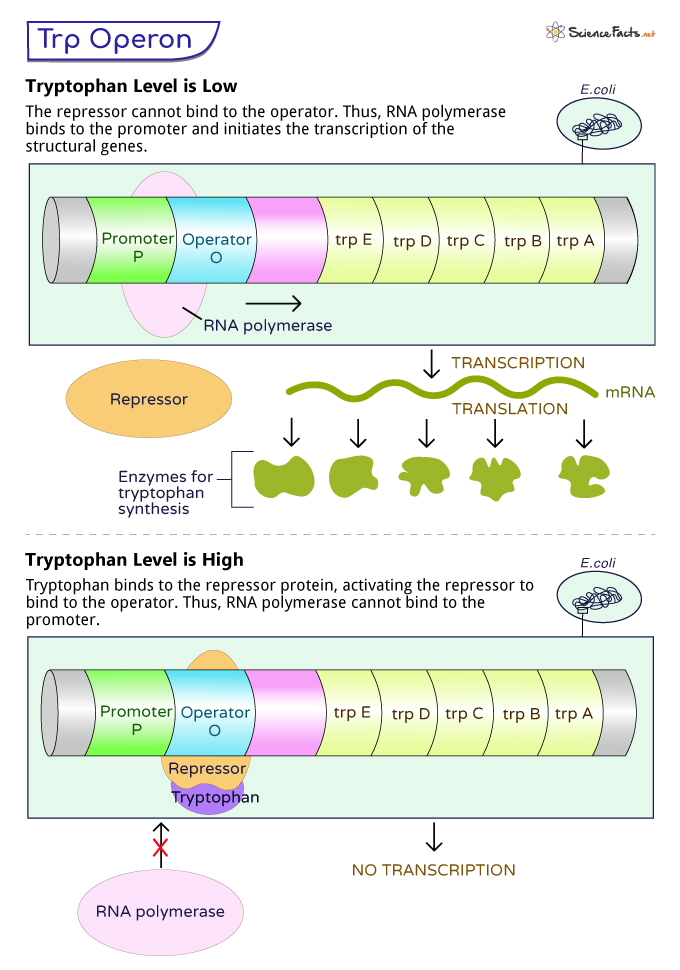
When the Tryptophan Level is Low
When the bacterial cell is in a tryptophan-deficient environment, it must produce tryptophan to support essential cellular processes. Under these conditions, the trp operon is activated to enable the synthesis of enzymes required for tryptophan biosynthesis.
In such a state, the repressor cannot bind to the operator of the trp operon, which makes way for the RNA polymerase to access the promoter region. Once RNA polymerase binds to the promoter, it initiates the transcription of the structural genes (trpE, trpD, trpC, trpB, and trpA) present in the trp operon. These structural genes produce the enzymes involved in tryptophan biosynthesis.
Ribosomes then translate the mRNA transcribed from the structural genes into functional enzymes. These enzymes work together to catalyze the sequential steps of tryptophan biosynthesis, starting from chorismate to form tryptophan eventually.
When the Tryptophan Level is High
When the bacterial cell has ample tryptophan, there is no need to produce more tryptophan. In this scenario, the trp operon is repressed to conserve cellular resources and prevent unnecessary tryptophan synthesis.
In such a situation, tryptophan molecules bind to the repressor protein (TrpR), inducing a conformational change in the protein. This activated repressor then attaches to the operator region of the trp operon. Once bound to the operator, it obstructs RNA polymerase from accessing the promoter region. It prevents RNA polymerase from initiating the transcription of the structural genes, thus preventing tryptophan biosynthesis.
Attenuation of the Trp Operon
In some bacterial species, an additional level of regulation called ‘attenuation’ exists in the trp operon. It involves an attenuator region located in the leader sequence of the mRNA transcribed from the trp operon.
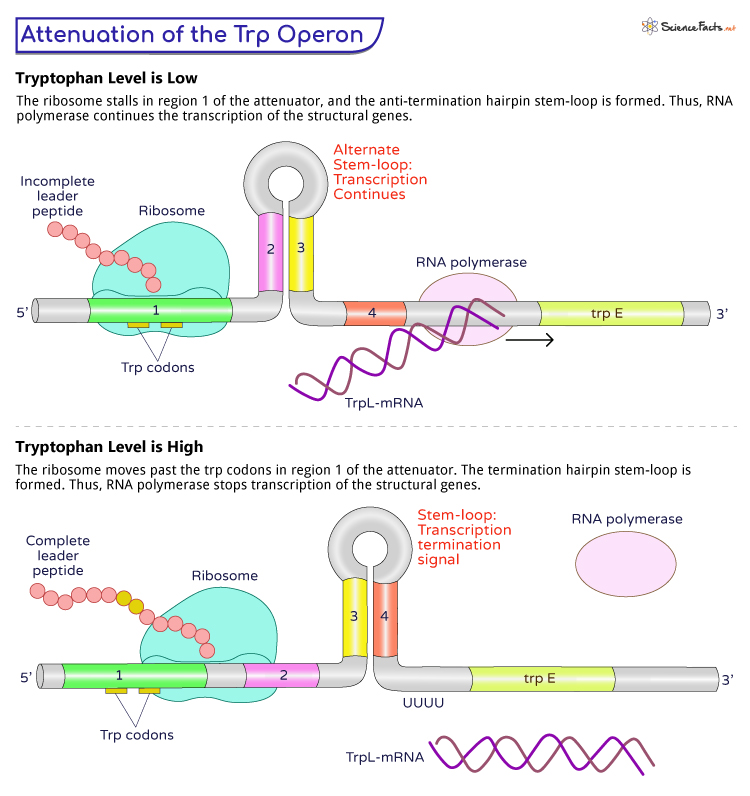
The attenuator region contains four distinct areas, designated as 1, 2, 3, and 4. Depending on the presence or absence of tryptophan in the cell, the mRNA transcribed from the attenuator region can fold into two alternative secondary structures.
During Low Levels of Tryptophan
Formation of Anti-termination Structure: In tryptophan-deficient conditions, the ribosome translating the mRNA encounters a sequence of consecutive tryptophan codons (trp codons) in region 1 of the attenuator. This ribosome stalling leads to the formation of an anti-termination hairpin in regions 2 and 3. This structure prevents the formation of a terminator hairpin in regions 3 and 4, allowing RNA polymerase to continue transcription into the structural genes. This results in the synthesis of enzymes necessary for tryptophan biosynthesis.
If tryptophan is abundant, a particular structure in the attenuator causes transcription to terminate before the structural genes are reached. In contrast, the mRNA forms a different structure in tryptophan-deficient conditions, allowing transcription to continue into the structural genes, promoting tryptophan synthesis.
During High Levels of Tryptophan
Formation of Termination Structure: When tryptophan is abundant in the cell, there is a sufficient supply of charged tRNAs (transfer RNAs) carrying tryptophan to the ribosome. The ribosome translating the mRNA moves past through the trp codons in region 1 without stalling. It leads to a termination hairpin in regions 2 and 3. The termination hairpin prompts the RNA polymerase to terminate transcription prematurely before reaching the structural genes. It prevents the synthesis of enzymes for tryptophan biosynthesis.
Thus the formation of the anti-termination or the termination structure in the attenuator region determines whether the trp operon is fully transcribed or terminated prematurely. This mechanism of fine-tuning the transcription based on tryptophan levels allows the bacterial cell to regulate tryptophan biosynthesis as required for its metabolic needs efficiently.
Combined Regulation of Attenuator and Repressor Proteins
The attenuation mechanism works with the repressor protein (TrpR) regulation in response to tryptophan levels. When tryptophan is abundant, the repressor binds to the operator region. It prevents RNA polymerase from accessing the promoter, leading to operon repression. At the same time, the attenuation mechanism ensures that even if transcription starts, it is prematurely terminated before the structural genes, further reducing tryptophan synthesis.
In contrast, when tryptophan is limited, the repressor is inactive, allowing RNA polymerase to bind to the promoter and initiate transcription. Simultaneously, the attenuation mechanism ensures that transcription proceeds smoothly into the structural genes, promoting tryptophan biosynthesis.
Trp Operon vs. Lac Operon
The key differences between the lac operon and trp operon are:
| Basis | Trp Operon | Lac Operon |
|---|---|---|
| 1. Function | Controls the synthesis of enzymes involved in tryptophan biosynthesis | Responsible for the metabolism of lactose, a disaccharide sugar. It is activated when lactose is present and glucose levels are low. |
| 2. Regulation | Regulated by negative control through the repressor protein (TrpR) and attenuation | Regulated negatively through the repressor protein |
| 3. Repressor Protein | Has a repressor protein (TrpR) that binds to the operator in the presence of tryptophan | Has a repressor protein (LacI) that binds to the operator without lactose |
| 4. Inducer Molecule | No inducer | Lactose acts as an inducer. |
| 5. Inducible/Repressible System | Repressible system | Inducible system |
| 6. Mode of Repression | Uses negative control (repressor binding to the operator) and attenuation | Done by the repressor protein (LacI) |
| 7. Feedback Regulation | Feedback regulation occurs | No feedback regulation occurs |
-
References
Article was last reviewed on Tuesday, August 29, 2023