Restriction Enzymes
Restriction enzymes, or restriction endonucleases, are enzymes initially isolated from bacteria that cleave DNA at sequence-specific sites, producing known DNA fragments. They do not discriminate between the DNA of bacteria, fungi, mice, or humans. If they recognize their target site, they cut.
These enzymes are widely found in bacteria and archaea, providing them with defense against invading bacteriophages. Inside a prokaryotic cell, the restriction enzymes selectively cut specific viral DNA sequences, a process known as restriction digestion. However, the host protects its DNA from such cleavage through methylation.
Werner Arber, Hamilton O. Smith, and Daniel Nathans discovered and characterized them in the late 1960s and early 1970s. A restriction enzyme is named after the genus, species, and strain of bacteria that produced it. For example, the enzyme EcoRI is produced by Escherichia coli strain RY13.
More than 3,600 restriction enzymes are known, of which over 3,000 are well characterized. Almost 800 such enzymes are commercially available for recombinant DNA technology and genetic engineering.
Types of Restriction Enzymes
Naturally occurring restriction enzymes are of 5 types based on several factors like the nature of the target sequence, the position of their cleavage site relative to their target sequence, the composition of the enzyme, and the requirement of cofactors.
Type I
Type I restriction enzymes’ cleavage sites are far away (at least 1000 base pairs) from their recognition site. They require ATP and the cofactor S-adenosyl-L-methionine to function.
Type II
Type II restriction enzymes are the most well-known and extensively used among all types. The cleavage sites of these enzymes are undivided and palindromic, with a length of 4–8 nucleotides. They require Mg2+ as a cofactor. These enzymes cleave the double helix’s phosphodiester bond, creating a staggered or blunt end.
Examples
Here is a list of common type II restriction enzymes used for molecular cloning with their recognition sites.
| Enzyme | Source | Recognition Site |
|---|---|---|
| EcoRI | Escherichia coli | 5’GAATTC3’CTTAAG |
| BamHI | Bacillus amyloliquefaciens | 5’GGATCC3’CCTAGG |
| HindIII | Haemophilus influenzae | 5’AAGCTT3’TTCGAA |
| NotI | Nocardia otitidis | 5’GCGGCCGC3’CGCCGGCG |
| SmaI | Serratia marcescens | 5’CCCGGG3’GGGCCC |
| PstI | Providencia stuartii | 5’CTGCAG3’GACGTC |
| KpnI | Klebsiella pneumoniae | 5’GGTACC3’CCATGG |
| AluI | Arthrobacter luteus | 5’AGCT3’TCGA |
| SacI | Streptomyces achromogenes | 5’GAGCTC3’CTCGAG |
| TaqI | Thermus aquaticus | 5’TCGA3’AGCT |
Type III
Type III restriction enzymes’ cleave sites are a short distance (about 20-30 base pairs) from their recognition site, which are two separate non-palindromic sequences, inversely oriented. They require ATP, while S-adenosyl-L-methionine acts as the stimulator but is not required to carry out the process.
Type IV
Type IV restriction enzymes recognize the modified, methylated DNA of E. coli.
Type V
Type V restriction enzymes use guide RNAs to target specific non-palindromic sequences found in invading organisms. They cut DNA of variable length provided with a suitable guide RNA.
How do Restriction Enzymes Work
Restriction enzymes bind to a specific sequence in the DNA, known as the restriction site. Each enzyme recognizes one or a few restriction sites specific to it.
After recognizing its target, the restriction enzyme binds and makes a double-stranded cut in the DNA molecule. The cut is typically created at or near the restriction site, making a staggered or blunt end depending on the restriction enzyme.
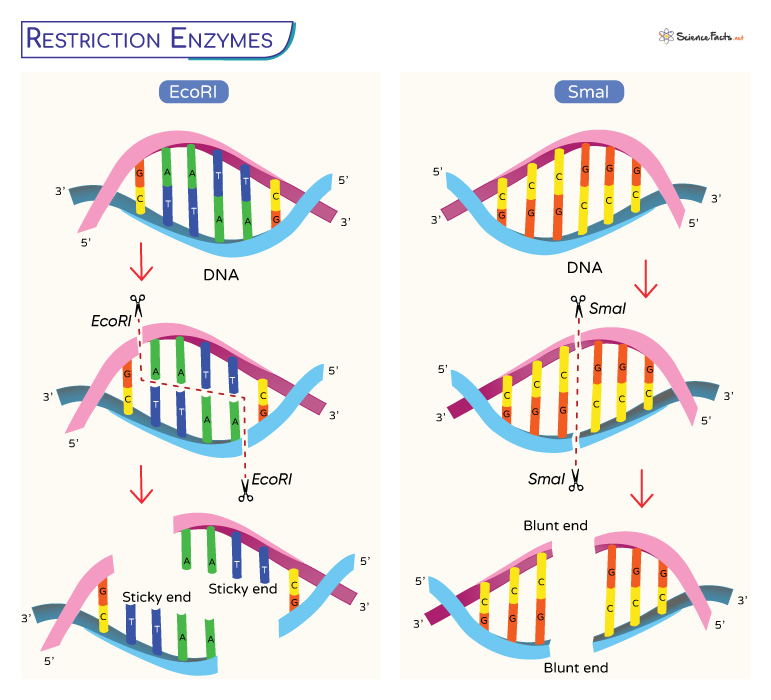
EcoRI and SmaI
Here, we will explain how the restriction enzymes recognize and cut at a DNA sequence with the help of the enzymes EcoRI and SmaI.
EcoRI recognizes and cuts within the sequence:
5’GAATTC
3’CTTAAG
The cut is precise, producing single-stranded sticky overhangs.
5′—GAGCT C—3′
3′—C TCGAG—5′
If another piece of DNA has matching overhangs (when cut by EcoRI), the complementary parts stick together by base pairing. Such overhangs are thus called sticky ends. They play a crucial role in cloning as they hold two pieces of DNA together so that they can be sealed by DNA ligase.
Not all restriction enzymes produce sticky ends. Some are blunt-cutters, which cut straight down the middle of a target sequence and have no overhang. Restriction enzyme SmaI is a blunt end cutter.
SmaI recognizes and cuts within the sequence:
5’CCCGGG
3’GGGCCC
It cuts at the target site and produces blunt end overhangs.
5′—CCC GGG—3′
3′—GGG CCC—5′
DNA ligase joins blunt ends through a covalent bond like sticky end ligation. However, blunt end ligation is less efficient as no single-stranded overhangs hold the DNA molecules in position.
What are Restriction Enzymes Used For
Restriction enzymes are essential and serve as a valuable tool in various scientific and biotechnological applications:
- Recombinant DNA Technology: Scientists use restriction enzymes to cut DNA at specific sites and insert new genetic material, such as genes with desired traits, into the gaps. This process helps create genetically modified organisms and has revolutionized fields like medicine and agriculture.
- DNA Fingerprinting: In forensic science, restriction enzymes create unique DNA profiles for individuals. Cutting DNA at specific recognition sites helps to separate and analyze the resulting fragments. These profiles are crucial in solving crimes and identifying individuals with unmatched accuracy.
- Genetic Mapping: Restriction enzymes assist in mapping the locations of genes on chromosomes. By cutting DNA at specific sites, researchers can analyze how genes are arranged and linked to particular traits or diseases.
- Cloning: In cloning, restriction enzymes help create identical copies of genes or organisms by cutting and inserting specific DNA fragments into host cells. After cutting with a restriction enzyme, the DNA fragment is inserted into a vector, often a plasmid or a viral genome. When the vector containing the DNA fragment is introduced into a host cell, it replicates with the host DNA, producing numerous identical copies of the inserted DNA.
- Genomic Library Construction: Creating genomic libraries is essential for large-scale genomic research. Researchers use restriction enzymes to fragment an organism’s DNA into pieces. These fragments are then inserted into vectors, creating a library of DNA fragments representing the entire genome. These libraries are used in the functional analysis of genes and finding evolutionary relatedness.
The study of restriction enzymes continues to evolve, with scientists discovering new enzymes and applications regularly. Thus, it is the future of genetic research.
-
References
Article was last reviewed on Friday, November 3, 2023