DNA Replication
DNA replication is the process through which a cell’s DNA forms two exact copies of itself. It occurs in all living organisms as it forms the basis of inheritance in all living organisms. The enzyme carrying out transcription is called DNA polymerase.
The primary mechanism of DNA replication is similar across all organisms. Here, we will focus on DNA replication as it takes place in the bacterium E. coli. However, the procedure is the same in humans and other eukaryotes.
Why is DNA Replication such an Important Process
DNA replication’s primary purpose is to enable living organisms to reproduce. The only way to replace the cells is to copy the cell’s information. It is what DNA replication does. After cell division by mitosis or meiosis, the two daughter cells must contain the same genetic information, or DNA, as the parent cell. Without the accurate copying of the DNA, life would cease to continue as existing organisms would not be able to reproduce.
Role of DNA Polymerase in DNA Replication
DNA polymerase is the main enzyme that carries out DNA replication. It adds nucleotides one at a time to the growing DNA strand. The nucleotides added are arranged such that they are complementary to the template.
Some key features of DNA polymerase are:
- Needs a template strand to copy
- Adds nucleotides (ATP, GTP, CTP) to the 3′ end of a DNA strand one at a time
- Requires a pre-existing chain or short stretch of nucleotides called an RNA primer
- Removes wrongly added nucleotides by proofreading the activity
- Adds 1,000 nucleotides per second in E. coli and 50 nucleotides per second in humans
In E. coli, two main DNA polymerases are involved in DNA replication: DNA pol III and DNA pol I.
When and Where does DNA Replication Occur
It happens in the S phase of cell division. DNA must replicate before cells divide so that the two resulting daughter cells contain the same genetic information as the parent cell.
DNA replication occurs in the nucleus of all eukaryotic cells and the nucleoid of prokaryotes.
How is DNA Replicated
Replication happens in three fundamental stages. They are described below in order:
Step 1: Initiation
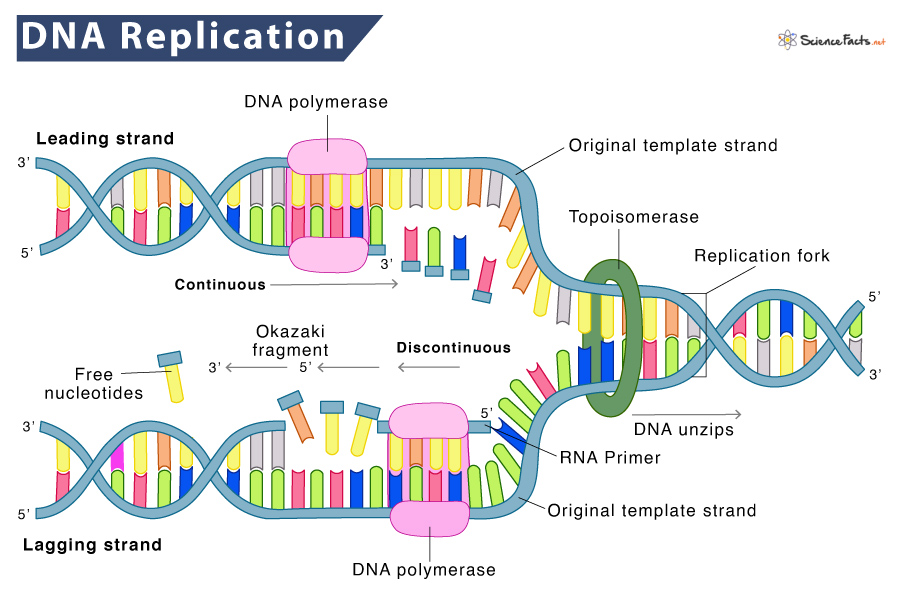
Unzipping of DNA
DNA replication starts at a particular location on the DNA, called the origin of replication. It is the region where the DNA is unzipped. They have a specific sequence covering about 245 base pairs, mostly A/T base pairs and fewer GT-base pairs. The fewer hydrogen bonds in the AT-rich sequence make the DNA strands separate easily.
Here, the initiator protein, helicase, recognize and then binds to the origin to unwind the DNA. When the DNA unwinds, two Y-shaped structures called replication forks are formed, together making up the replication bubble. The two forks move in opposite directions as the replication proceeds. Single-strand binding (SSB) proteins coat the separated strands of DNA to prevent them from rewinding into a double helix.
Primer Synthesis by Primase
Meanwhile, as the helicase separates the strands, a different enzyme called primase attaches to each strand. It synthesizes a short stretch of nucleotides complementary to the template called an RNA primer. It marks the beginning of the polymerization of the DNA strand.
Step 2: Elongation
After the primer is synthesized, DNA polymerase III adds nucleotides with its 5′-phosphate group to the 3′ end of the last nucleotide of the primer. Synthesis of the growing strand in the 5′-to-3′ direction involves adding nucleotides in a complementary order to the template strand. According to the Watson-Crick DNA model, adenine always pairs with thymine, and cytosine pairs with guanine. This process is known as complementary base pairing.
DNA Polymerase Can Move in One Direction
Two molecules of DNA polymerase III at the replication fork carry out replication. However, DNA polymerases can only make DNA in the 5′ to 3′ direction. It poses a problem. One original strand runs in the 5′ to 3′ direction and the other in the 3′ to 5′ direction. Thus, the two new strands need to be replicated slightly differently.
Synthesis of Leading and Lagging Strand
The strand that runs 5′ to 3′ in the direction of the replication fork is easily replicated continuously as the DNA polymerase moves in the same direction as the replication fork. This strand is called the leading strand.
The replication of the other strand, which runs in the 5′ to 3′ direction away from the fork, is made discontinuously. It happens because as the fork moves forward, the DNA polymerase (which is moving away from the fork) comes off and then reattach to the newly exposed DNA. This strand is called the lagging strand. The small fragments are called Okazaki fragments, named after the discoverer, Japanese molecular biologist Reiji Okazaki, who discovered them. Thus, the leading strand can be extended from one primer alone. In contrast, the lagging strand needs a new primer for each short Okazaki fragment.
Role of Other Proteins
Some other proteins, in addition to helicase, DNA polymerase, and primase, are also involved during the process to ensure that elongation can occur smoothly.
One such protein is the sliding clamp, which holds DNA polymerase III in place as they synthesize DNA. It helps keep the DNA polymerase in the lagging strand from dislocation when it restarts at a new Okazaki fragment.
The other one is the Topoisomerase, which prevents the DNA double helix ahead of the replication fork from tight wounding as the helicase help to open up the DNA. It makes temporary nicks in the helix to release the tension and then reseals the nicks to errors in replication.
Step 3: Termination
Removal of RNA Primers by Exonuclease
Once the continuous and discontinuous strand synthesis is complete, an exonuclease removes all RNA primers in the lagging strand. They are then replaced with DNA through the activity of DNA polymerase I, the other polymerase involved in the replication. The nicks that remain after the primers are replaced get sealed by the enzyme DNA ligase forming a complete strand.
The Role of Tus Protein
Since bacterial chromosomes are circular, replicated ends when the two replication forks meet on the opposite end of the replicated DNA. This step is regulated by a specific termination sequence that, when bound by the Tus protein, enables only one replication fork to pass through while preventing the other. As a result, the replication forks meet within the termination region, causing dissociation of the polymerase and release of the newly synthesized strands.
Replication in Eukaryotes
Prokaryotic and eukaryotic replications are dissimilar in many ways, although they follow the same pattern.
Similarities
- Require primers for initiation
- DNA elongation occurs in the 5′ to 3′ direction
- Replication is continuous in the leading strand and discontinuous in the lagging strand.
- However, there are also some differences between them.
Differences
- Eukaryotic replication has many replication origins (often thousands). Unlike prokaryotes, the origin is not conserved and varies according to the species.
- Eukaryotic replication utilizes five different DNA polymerases – α, β, γ, δ, and ε.
- The DNA in eukaryotes is wound around proteins called histones, and thus, unwinding and rewinding happen in a different way
- In bacteria with circular DNA, nucleotides quickly replace the excised primer, leaving no gap in the newly synthesized DNA. However, linear eukaryotic DNA has a small gap left at the end of the DNA due to the lack of a 3′-OH group for nucleotides to bind. A unique enzyme called telomerase prevents the DNA molecule from getting shorter in every round of DNA replication
- Compared to prokaryotes, DNA polymerase adds nucleotides at a much slower rate of 50 nucleotides per second
Is DNA Replication Semi-conservative
The result of DNA replication is two new DNA molecules. Each DNA consists of one newly synthesized strand while the other is obtained from the parent molecule. Thus, DNA replication is semi-conservative.
-
References
Article was last reviewed on Friday, February 17, 2023